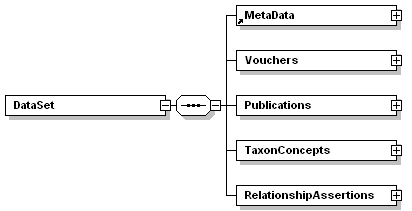
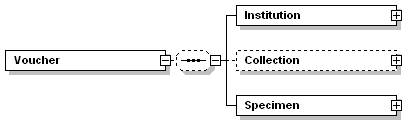
| annotation |
| documentation | The date and time expressed in a way conforming to a subset of ISO 8601 | | appInfo | <sea:FullName xml:lang="en">ISO/ANSI Date and Time</sea:FullName>
<sea:Content xml:lang="en">A representation of the date and time of (the beginning of) an event in ISO/ANSI standard structured format.</sea:Content>
<sea:Audience>BioCASE</sea:Audience>
<sea:Audience>CODATA TDWG</sea:Audience>
<sea:Audience>HISPID</sea:Audience>
<sea:Reviewer>M. Doering, A. Guentsch and W. Berendsohn 2003</sea:Reviewer>
<sea:ExistingStandard>ISO Standard 8601:1988</sea:ExistingStandard>
<sea:ExistingStandard>HISPID</sea:ExistingStandard>
<sea:ExistingStandard>ITF</sea:ExistingStandard>
<sea:Content xml:lang="en">Any chronologically acceptable date and time in ISO/ANSI format</sea:Content>
<sea:Example xml:lang="en">1966-12-05T23:55:46</sea:Example>
<sea:Example xml:lang="en">1925-02-21</sea:Example>
<sea:Example xml:lang="en">1925-02</sea:Example>
<sea:Example xml:lang="en">1925</sea:Example>
<sea:Example xml:lang="en">--12-05T23:55:46</sea:Example>
<sea:Example xml:lang="en">---05T23:55</sea:Example>
<sea:Example xml:lang="en">1966-12-05T23:55</sea:Example>
<sea:Rule xml:lang="en">This field is a representation of the date and time in its structured form</sea:Rule>
<sea:Rule xml:lang="en">The values are in order Year-Month-Day, separated by hyphens, followed by the letter T and the time, in order Hour:Minutes:Seconds, separated by colons</sea:Rule>
<sea:Rule xml:lang="en">Partial dates in the form of Year-Month or Year only are acceptable</sea:Rule>
<sea:Rule xml:lang="en">The year is expressed as a 4-digit value, left zero padded if necessary</sea:Rule>
<sea:Rule xml:lang="en">The year can be omitted, the month is than preceeded by two hyphens</sea:Rule>
<sea:Rule xml:lang="en">The year and the month can be omitted, the day is than preceeded by three hyphens</sea:Rule>
<sea:Rule xml:lang="en">The month of the year is expressed in as 2-digit value, left zero padded if necessary, ranging between 01 and 12</sea:Rule>
<sea:Rule xml:lang="en">The day of the month is expressed as a 2-digit value, left zero padded if necessary, ranging between 01 and 31.</sea:Rule>
<sea:Rule xml:lang="en">Parts can be omitted from the right, whereas a year is replaced by '--' and only a day would look like '---31'.</sea:Rule>
<sea:Rule xml:lang="en">The hour is expressed as a 2-digit value, left zero padded if necessary, ranging between 01 and 31.</sea:Rule>
<sea:Comment xml:lang="en">This field will nearly always be a representation derived from the date on the specimen. The DateText field is available for recording verbatim date information for generalized or ambiguous dates.</sea:Comment>
<sea:Comment xml:lang="en">The year must be represented as four digits to avoid ambiguity between centuries. Month and Day values are left zero padded to two digits to facilitate machine readability.</sea:Comment>
<sea:Comment xml:lang="en">In the rare instances of multiple exact dates or periods those can be recorded in this DateText field.</sea:Comment>
<sea:Comment xml:lang="en">In the rare instances of dates for periods before the year 0000, BC and similar dates can be recorded in this DateText field.</sea:Comment>
<sea:Comment xml:lang="en">Parts can be omitted from the right, whereas a year is replaced by '--' and only a day would look like '---31'.</sea:Comment>
<sea:Comment xml:lang="en">In the ISO standard element, time can only be specified when there is a full date.</sea:Comment> |
|
| source |
<xs:simpleType name="DateTimeISOType1">
<xs:annotation>
<xs:documentation xml:lang="en">The date and time expressed in a way conforming to a subset of ISO 8601</xs:documentation>
<xs:appinfo>
<sea:FullName xml:lang="en">ISO/ANSI Date and Time</sea:FullName>
<sea:Content xml:lang="en">A representation of the date and time of (the beginning of) an event in ISO/ANSI standard structured format.</sea:Content>
<sea:Audience>BioCASE</sea:Audience>
<sea:Audience>CODATA TDWG</sea:Audience>
<sea:Audience>HISPID</sea:Audience>
<sea:Reviewer>M. Doering, A. Guentsch and W. Berendsohn 2003</sea:Reviewer>
<sea:ExistingStandard>ISO Standard 8601:1988</sea:ExistingStandard>
<sea:ExistingStandard>HISPID</sea:ExistingStandard>
<sea:ExistingStandard>ITF</sea:ExistingStandard>
<sea:Content xml:lang="en">Any chronologically acceptable date and time in ISO/ANSI format</sea:Content>
<sea:Example xml:lang="en">1966-12-05T23:55:46</sea:Example>
<sea:Example xml:lang="en">1925-02-21</sea:Example>
<sea:Example xml:lang="en">1925-02</sea:Example>
<sea:Example xml:lang="en">1925</sea:Example>
<sea:Example xml:lang="en">--12-05T23:55:46</sea:Example>
<sea:Example xml:lang="en">---05T23:55</sea:Example>
<sea:Example xml:lang="en">1966-12-05T23:55</sea:Example>
<sea:Rule xml:lang="en">This field is a representation of the date and time in its structured form</sea:Rule>
<sea:Rule xml:lang="en">The values are in order Year-Month-Day, separated by hyphens, followed by the letter T and the time, in order Hour:Minutes:Seconds, separated by colons</sea:Rule>
<sea:Rule xml:lang="en">Partial dates in the form of Year-Month or Year only are acceptable</sea:Rule>
<sea:Rule xml:lang="en">The year is expressed as a 4-digit value, left zero padded if necessary</sea:Rule>
<sea:Rule xml:lang="en">The year can be omitted, the month is than preceeded by two hyphens</sea:Rule>
<sea:Rule xml:lang="en">The year and the month can be omitted, the day is than preceeded by three hyphens</sea:Rule>
<sea:Rule xml:lang="en">The month of the year is expressed in as 2-digit value, left zero padded if necessary, ranging between 01 and 12</sea:Rule>
<sea:Rule xml:lang="en">The day of the month is expressed as a 2-digit value, left zero padded if necessary, ranging between 01 and 31.</sea:Rule>
<sea:Rule xml:lang="en">Parts can be omitted from the right, whereas a year is replaced by '--' and only a day would look like '---31'.</sea:Rule>
<sea:Rule xml:lang="en">The hour is expressed as a 2-digit value, left zero padded if necessary, ranging between 01 and 31.</sea:Rule>
<sea:Comment xml:lang="en">This field will nearly always be a representation derived from the date on the specimen. The DateText field is available for recording verbatim date information for generalized or ambiguous dates.</sea:Comment>
<sea:Comment xml:lang="en">The year must be represented as four digits to avoid ambiguity between centuries. Month and Day values are left zero padded to two digits to facilitate machine readability.</sea:Comment>
<sea:Comment xml:lang="en">In the rare instances of multiple exact dates or periods those can be recorded in this DateText field.</sea:Comment>
<sea:Comment xml:lang="en">In the rare instances of dates for periods before the year 0000, BC and similar dates can be recorded in this DateText field.</sea:Comment>
<sea:Comment xml:lang="en">Parts can be omitted from the right, whereas a year is replaced by '--' and only a day would look like '---31'.</sea:Comment>
<sea:Comment xml:lang="en">In the ISO standard element, time can only be specified when there is a full date.</sea:Comment>
</xs:appinfo>
</xs:annotation>
<xs:restriction base="xs:string">
<xs:pattern value="\d\d\d\d(\-(0[1-9]|1[012])(\-((0[1-9])|1\d|2\d|3[01])( (0\d|1\d|2[0-3])(:[0-5]\d){0,2})?)?)?|\-\-(0[1-9]|1[012])(\-(0[1-9]|1\d|2\d|3[01]))?|\-\-\-(0[1-9]|1\d|2\d|3[01])"/>
</xs:restriction>
</xs:simpleType> |